Code: https://github.com/gusmmm/demo_basic_structured_output
A hands-on demonstration of how AI can transform unstructured medical texts into structured, analyzable data
—
The Challenge We Face
As physicians, we write countless clinical notes, discharge summaries, and case reports. These narratives contain invaluable diagnostic information, but extracting this data systematically for research, quality improvement, or clinical decision support remains a significant challenge.
Traditional methods – manual chart review or basic keyword searching – are time-consuming, error-prone, and don’t scale.
What if we could automatically extract structured diagnostic information from our clinical texts with high accuracy? This article demonstrates a practical solution using AI that any physician with basic Python knowledge can implement and customize.
What This Tool Does
The demonstration repository presents a focused solution: extracting medical diagnoses from clinical narratives and outputting structured JSON data. Here’s what happens:
1. Input Unstructured clinical text (discharge summary, case report, clinical note)
2. Processing AI-powered extraction using Google’s Gemini model
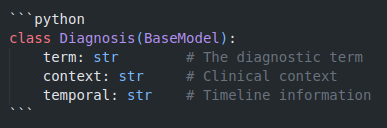
3. Output Structured JSON containing each diagnosis with its context and temporal information
Example Transformation
Input text: A 59-year-old male patient was diagnosed with ST-segment elevation myocardial infarction on an electrocardiogram (ECG). During preparation for coronary angiography, repeated cardiac arrest occurred.
Structured output:

When This Approach Is Valuable
Immediate Applications
Research Data Extraction Quickly extract diagnostic patterns from case series or retrospective chart reviews. Instead of manually coding hundreds of discharge summaries, process them automatically and focus on analysis.
Quality Improvement Projects Extract complications, adverse events, or specific diagnoses across patient populations to identify trends and improvement opportunities.
Clinical Decision Support Build databases of diagnostic patterns to support differential diagnosis or identify missed diagnoses in similar presentations.
Broader Use Cases
This same approach can extract:
- Medications and dosages from clinical notes
- Procedures and their outcomes from operative reports
- Symptoms and their severity from emergency department notes
- Laboratory values and their interpretations from progress notes
- Family history patterns from admission histories
- Social determinants of health from social work evaluations
Technical Implementation: Simple but Powerful
The solution uses three key components that make it both accessible and robust:
1. Text Preprocessing
Cleans and normalizes clinical text while preserving medical context—removing artifacts from copy-paste operations, standardizing formatting, and ensuring consistent input for AI processing.
2. AI-Powered Extraction
Leverages Google’s Gemini 2.5 Flash model with structured output capabilities. The AI understands medical terminology, context, and relationships without requiring extensive training on medical-specific datasets.
3. Pydantic Data Models
Ensures extracted data follows a consistent schema, providing type safety and validation. This prevents malformed outputs and ensures downstream compatibility.
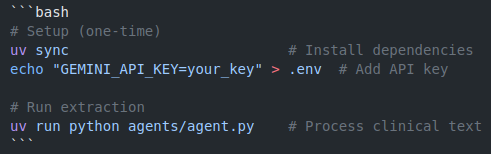
Running the Demo
The implementation is straightforward for any physician comfortable with basic Python:
The tool processes the sample case (a complex ICU patient with multiple diagnoses) and extracts 10 distinct diagnostic terms with their clinical context and temporal relationships.
Beyond the Demo: Production-Ready Clinical Informatics
While this demonstration shows the core extraction capability, implementing this in a clinical environment requires several additional steps to make the data truly useful and interoperable:
1. Terminology Mapping and Standardization
SNOMED-CT Integration: Raw extracted terms must be mapped to standardized medical terminology. SNOMED-CT provides the most comprehensive clinical terminology system globally.
Implementation considerations:
- Use SNOMED-CT’s Python APIs or UMLS Metathesaurus
- Implement fuzzy matching for term variations
- Handle synonyms and alternative expressions
- Validate mappings with clinical experts
- Maintain version control for terminology updates
2. HL7 FHIR for Interoperability
Transform extracted data into HL7 FHIR resources for good integration with electronic health records and other clinical systems.
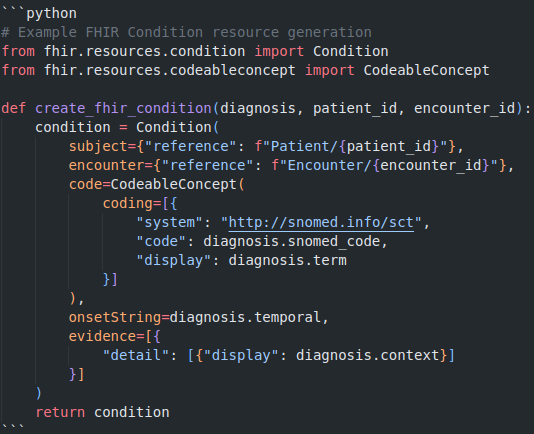
3. Database Architecture for Clinical Data
- PostgreSQL Implementation (recommended for structured clinical data):
- MongoDB Alternative (for semi-structured clinical data)
4. Clinical Validation Workflow
Human-in-the-Loop Validation:
- Implement review queues for extracted diagnoses
- Provide clinician interfaces for validation/correction
- Track inter-rater reliability metrics
- Implement feedback loops to improve extraction accuracy
Automated Quality Checks
5. Integration with Clinical Workflows
EHR Integration Points
- Real-time processing of new clinical notes
- Batch processing of historical data
- Integration with clinical decision support systems
- Quality metric dashboards for clinical leadership
API Development
The Path Forward
This demonstration represents a basilar approach to clinical text processing that can transform how we extract value from clinical narratives. While the core extraction is remarkably straightforward with AI, the real work lies in building the infrastructure for clinical-grade data management, validation, and integration.
The combination of AI-powered extraction, standardized terminologies like SNOMED-CT, interoperable formats like HL7 FHIR, and robust database architectures creates a pathway from clinical narratives to actionable clinical intelligence.
For MDs interested in clinical informatics, this represents an accessible entry point into AI-powered clinical data extraction. The technical barriers are lower than ever, and the potential impact on clinical research, quality improvement, and patient care is substantial.
The complete working demonstration is available in the accompanying repository, ready for clinical informaticists and physician-developers to adapt for their specific use cases.
—
## Technical Requirements Summary
- Python 3.12+ with UV package manager
- Google Gemini API (free tier available)
- SNOMED-CT license (free for many academic/research uses)
- Database PostgreSQL or MongoDB
- FHIR libraries Python FHIR libraries for interoperability
- Clinical validation Web interface for clinician review
The investment in time and infrastructure is modest compared to the potential for transforming clinical data into actionable insights.